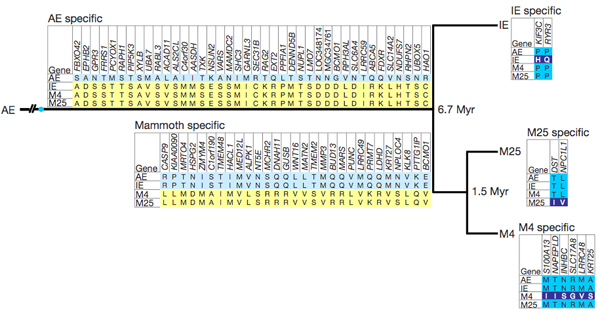
|
What is a SAP (Single Amino-acid Polymorphism)? We refer to an amino acid in a
woolly mammoth protein that differs from the corresponding (orthologous) elephant amino acid
(in any living elephant species) as a SAP, standing for single amino-acid polymophism.
Identifying functionally important SAPs has
been one of the major reasons for sequencing the woolly mammoth. To
start looking for them, we combined computational searches (designed to
enrich for validity and functional importance) with conventional PCR
amplification and Sanger sequencing of M4, M25,
African savanna elephant (Loxodonta africana) and
Indian elephant (Elephas maximus). Experimental confirmation of SAPs
is currently necessary because the rate of mammoth-elephant differences is about
the same as the rate of sequencing errors in individual reads.
Most likely, only a small fraction
the SAPs thus identified have
functional consequences. So far, we have investigated 351 predicted
SAPs between mammoth and elephant. For 260 of these, PCR products could
be obtained. These SAPs were further subjected to manual screening
for undesirable attributes such as lack of conservation (notably homoplasy)
at the SAP position, potential confusion with paralogs,
processed and unprocessed pseudogenes, and tandem or other duplicative
debris. The remaining 79 SAPs could be assigned to branches of the evolutionary tree
(AE = African elephant; IE = Indian elephant).
The number of SAPs assigned to the IE and M25
branches in the following picture is disproportionately small because
of the way we made our initial predictions (namely as AE/M4 SAPs). |
Figure
1: Experimentally verified amino-acid differences among African elephant,
Indian elephant, M4, and M25. |
 |